Introduction
I've always been fascinated by the idea of extending human lifespan. While the concept of uploading consciousness to a machine doesn't appeal to me, I'm drawn to the biological approach of reversing the aging process. After reading "Lifespan" by David Sinclair and "Ageless" by Andrew Steele , I was encouraged by the growing number of researchers working in this field, though the timeline remains scarily far away. Imagine being the generation who starts to die off just as they invent the cure for ageing!
Wanting to contribute somehow, but not yet having a background in biology, I thought I'd tackle this through something I do know. At the 2022 Blender Conference, I attended a presentation by Brady Johnston about Molecular Nodes, a Blender extension that allows the visualisation of proteins within Blender, using the geometry nodes feature.
Beginning this project I don't really have any prior knowledge in biology, save for what I did in school during my Biology GCSE so I'll likely be exploring tangents I find interesting along the way.
Objectives
To give me some direction as I work in this area I've decided to set some goals for myself. They're pretty flexible and I don't want to fence myself in too heavily, though I feel these three will point me in the right direction:
- Get more familiar with the science behind molecular biology
- Build Molecular Nodes locally and contribute something to the project
- Create a website to create interactive visualisations of any given protein
These are my initial goals, though at the moment there are a lot of unknown unknowns. It could turn out that I don't end up using molecular nodes and Blender for the final site I build. We'll have to wait and see!
Implementation
Creating a viewer
The first thing I wanted to do was to launch molecular nodes, export a glb and get it displayed on this page. The first step would be to create a 3D model viewer that I could reuse all over this site - you can see this below displaying the classic Blender monkey Suzanne
Loading 3D Model... 0%
Drag to rotate • Scroll to zoom • Right-click to pan
Wow, look at that! A viewer created using threejs displaying the Blender monkey Suzanne. That was implemented mostly by asking the Cursor agent to create a model viewer component for me. The first shot was pretty good, just needed some slight lighting tweaking and adding the ability to add a caption. Easy! Now we have a reusable component to use for visualising 3D models. It doesn't do anything too fancy just yet, though it does provide basic controls. Further down the line I might add drei to help with that, but for now let's get onto downloading some proteins!
Identifying a protein
It's a funny idea, downloading a protein. Thankfully there's a really handy repository of all known proteins called the Protein Data Bank (PDB), this is a global repo that contains information on the 3D structure of all known proteins. It's been going for over 50 years and hopefully should be an easy way to get the information required to visualise a protein molecule.
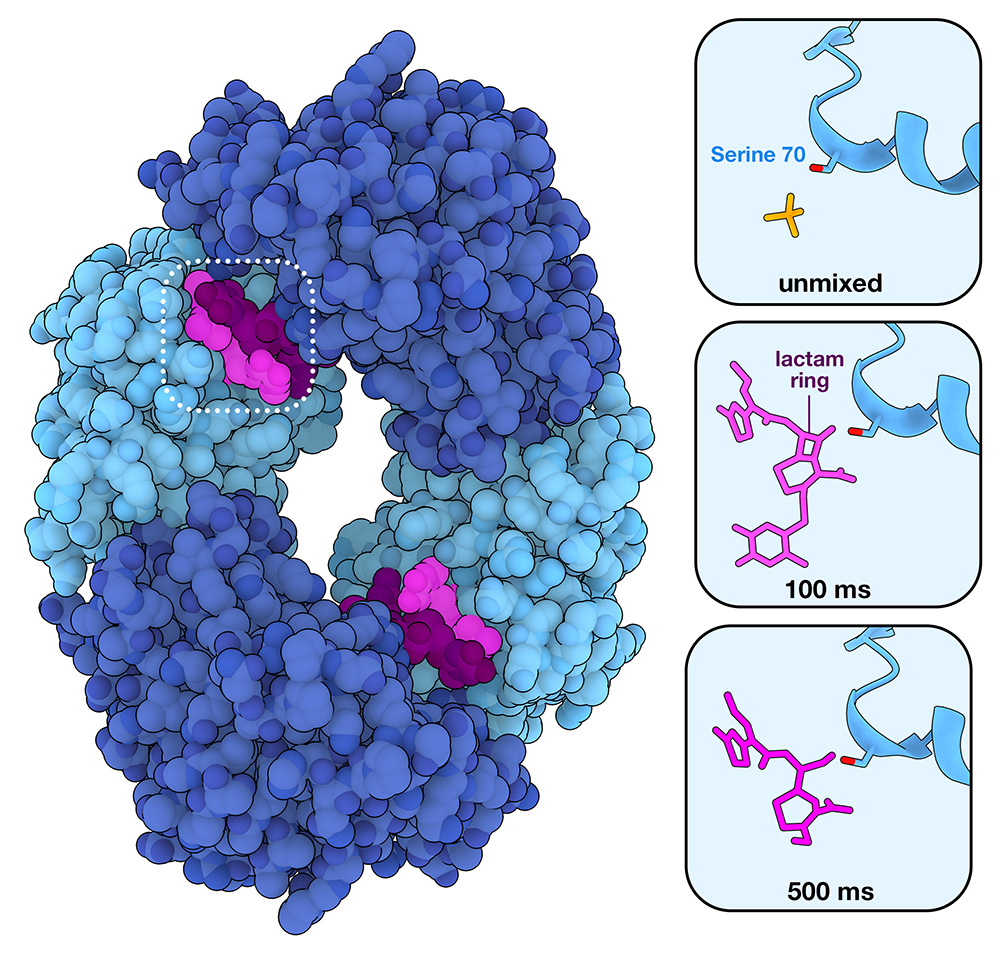
To begin I thought I'd look into the current molecule of the month - at the time of writing this is Beta-Lactamase. The name means very little to me at the moment, though it's got to be pretty important as 75% of antibiotics we use today are based on it. Here's the image from the summary page, it's a lovely toon-shader view of a molecule, though i'm no closer to knowing what a Serine 70 or a lactam ring is.
Looking into how the PDB works, it turns out that every protein that is discovered is given a unique identifier in format of a digit followed by 3 letters, case insensitive. As a new protein is discovered and submitted to the PDB, it's given an sequential identifier incrementing the characters, then eventually the numbers. For example, if 1AAA was the identifier of the first protein then the next submitted protein would be given 1AAB. Human haemoglobin that was figured out a while back is 1HHO, though the more recent coronavirus structure is 7PZK.
Anyway, back to our new favourite protein - Beta-Lactamase. It has a unique identifier of 1XPB and a lot of complicated statistics on its PDB page about how the structure was identified and how many atoms are in it (2,167) and how much it weighs (the same as 29,040 hydrogen atoms). I don't really care about this for the minute though - I want to know how those 2,167 atoms are represented in data.
It's worth noting that the PDB page did have a 3D protein viewer, as does every protein in the PDB. I chose to ignore this entirely as I wanted to approach this project from first principles without getting a sneak peak at what it was I wanted to do.
Downloading a protein
The PDB has a handy button to download the associated files - my first thought was "wow that's easy!". Unfortunately not. Clicking this button opens a drop down with all kinds of file formats that I've never heard of before: PDBx, mmCIF, PDBML/XML, 2fo-fc coefficients, biological assembly files to name a few.
I thought I had a pretty good handle on the 3D model formats out there, having messed around with both a bunch of CAD formats and mesh representation formats for work such as the gorgeous gLTF format format. I'd never even heard of any of these weird biological formats before though! I thought I'd just download the first one in the list, the 1XPB.FASTA file and open it up. This is what I saw:
HPETLVKVKDAEDQLGARVGYIELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRVDAGQEQLGRRIHYSQNDLVEYSPVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRLDRWEPELNEAIPNDERDTTMPAAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPLLRSALPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIAEIGASLIKHW
What an absolutely tiny file to represent a protein at 318 bytes. I thought I knew how to optimise 3D assets for the web through tools such as Draco , though that rarely gets anything below a few kilobytes.
Some brief research had me come across something I've heard of before - protein folding. While the .FASTA file contains the primary sequence of amino acids (the building blocks of a protein), it doesn't tell you how they are folded in 3D space and therefore has no info about atom coordinates or bond angles. Thankfully, a few years back some clever researchers at Google DeepMind figured out how to predict the structure of a protein soley from it's amino acids. The tool they developed is called AlphaFold and is really interesting to poke about with. There exists a AlphaFold page on Beta-Lactamase. At the top of this page are 3 download links: PDB file, mmCIF file and Predicted aligned error.
Given that AlphaFold displayed these 3 files for download had me thinking that the PBD and mmCIF formats would have the information needed to visualise a protein in 3D. I downloaded the files and immediately noticed they had hundreds of kilobytes of information - this had me thinking I was on the right track. A quick bit of research and I found out that the PDB format was developed in the 1970s (wow!) while the mmCIF was the more modern version developed in the 1990s that handles complex data a bit easier. I also found out that mmCIF stands for "macromolecular Crystallographic Info File" - I don't know what that means yet so I'll just keep referring to it as either mmCIF or the .cif format for now.
Exploring Molecular Node
Stepping back a moment, now I've learnt a little bit about how proteins have their information stored, I'm going to step sideways a bit and create my first proper protein visualisation. As mentioned, I know a tool called molecular nodes exists already. A long while back I opened it up and imported a protein, following the guide exactly without much understanding of what it was actually doing. So, first things first let's load in the 1XPB protein and see if we can get it looking lovely.
I opened up Blender 4.4, enabled the molecular nodes addon and proceeded to look at the sidebar it provided. Going into the scene tab there's a handy little field to add a PDB id, a way to select the file format (I chose .cif) and a button called fetch. Once I'd entered X1PD and hit fetch this beautiful molecule was loaded into my scene:
What a gorgeous assortment of spheres and connections. If I were a bacteria I'd for sure be scared of it!
Getting down to business, let's look at the actual stats of this molecule. To load it in added 2,033 objects to the scene using 65,862 vertices (the same kind of size as a mid poly game asset). Quite a lot going on there, significantly more than the small string of characters we saw above in the .FASTA file.
While cool, this isn't an optimal way to visualise the geometry in the browser. Right now there's a bunch of geometry being created and while we can instance it, I really want to play around with a particle system. To do this, I'll need to set up something that can manage a particle system.
Creating ParticularProteins
I decided to call this app Particular Proteins as it'll show a specific (or particular) protein in a particle system. Hilarious I know.
To do this I created a fresh new repo and set up a basic react app in it. For this I'm going to hop on the bandwagon of using shadcn.
This seemed like a nice time to set some subgoals for this project:
- Create a stunning react app with shadcn
- Host it on a subdomain of jamesledger.co.uk
- Either hook into a PDB api or host the data myself
- Display the pdb data in my app
It was easy enough to get set up with shadcn, though having not used tailwind in a little while it took me a moment to get back into it. The new registry setup offers some really cool ui elements, though I wanted to keep it basic for the moment and just made a simple card with an input form.
Hooking up to proteins.jamesledger.co.uk was easy enough too. I created a new AWS Amplify app and pointed it at the repo URL. Other than having to create and configure an amplify.yml file to allow the usage of pnpm this was easy too. Given how easy this was to do I think i'll for sure use this subdomain approach for side projects going forwards rather than spending a fortune on domains.
Here's a sneak peek of what it looks like at the moment.

At the moment I'm obsessed with the Flexoki colour scheme so I just copied over the setup I have for the main blog including the main flexoki.css. Later on i'll customise it a bit.
Using a PDB api was a little tricker. There's another handy page full of them, though as I'm not a biologist just yet I don't know what the majority of it actually means. I thought I'd start by taking a step back to the FASTA file format - that was something easier to parse. My thinking is that I'll set that up as the first slice and deal with the .cif file stuff later on.
Getting a FASTA file
The first thing I did was search "FASTA file api". This brought me to a handy PDB page that allows you to download specific FASTA files based on their PDB id. Perfect!
Also on this page was a button that said "Download a file containing sequences in FASTA format for all entries in the PDB archive". This was exactly what I was after! After all, it'd be a nice chance to poke about with databases. Downloading and unzipping this file revealed it was 343mb in size. Those biologists have certainly been very busy decoding proteins! The sheer size of this made me a bit more certain that I'd like to find an API rather than storing it myself. Doing it this way would also mean that it'd be kept up to date and I wouldn't have to worry about versioning.
After trying to figure out how the page obtained the FASTA file in the first place I ended up finding the RCSB's FASTA endpoints that had the following structure:
https://www.rcsb.org/FASTA/entry/{pdb_id}/display
Running a GET request against https://www.rcsb.org/FASTA/entry/1XPB/display gave me exactly what I was after, the raw contents of the FASTA file as seen below:
>1XPB_1|Chain A|BETA-LACTAMASE|Escherichia coli (562)
HPETLVKVKDAEDQLGARVGYIELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRVDAGQEQLGRRIHYSQNDLVEYSPVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRLDRWEPELNEAIPNDERDTTMPAAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPLLRSALPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIAEIGASLIKHW
Almost too easy - this was the same value we had before when downloading the file manually, only with an extra line of information. This extra info turned out to be:
- 1XPB_1 - The PDB id of the protein
- Chain A - The chain label for the entity in the structure
- BETA-LACTAMASE - The macromolecule/entity name
- Escherichia coli - The source organism with its NCBI taxonomy ID.
Interesting set of data there, I quite liked that it's from Escherichia coli - the government name of the E.Coli bateria. I remember growing this in a biology class once. After reading into it I'm still not 100% sure what chains and macromolecules are about, though i'll just leave it there for the moment and continue with the data visualisation.
Visualising the FASTA file
It was at this point I realised that I still hadn't figured out how to fold proteins the way that alphafold does. Due to this I'm skipping ahead to visualising a mmCIF file instead.
Visualising a mmCIF file
If you want to have a poke about with a mmCIF file click here to give it a download. For those who don't have all day, here's a snippet of the first few lines
data_1XPB
#
_entry.id 1XPB
#
_audit_conform.dict_name mmcif_pdbx.dic
_audit_conform.dict_version 5.397
_audit_conform.dict_location http://mmcif.pdb.org/dictionaries/ascii/mmcif_pdbx.dic
#
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
Yeah. A lot of metadata going on there, even spotted the 'loop_' so it seems like there's some kind of logic going on within this file format too, potentially as an optimisation. I'm hoping that some of those numbers are coordinates.
As I scrolled through the file I knew I'd found what I was looking for when I found the following lines
...
_atom_site.Cartn_x
_atom_site.Cartn_y
_atom_site.Cartn_z
...
Cartesian coordinates for the x,y,z axis of given atoms! These lines were about halfway through the file, all lines after that followed this structure:
ATOM 1 N N . HIS A 1 1 ? 18.376 1.585 34.443 1.00 21.34 ? 26 HIS A N 1
ATOM 2 C CA . HIS A 1 1 ? 18.239 1.879 35.857 1.00 19.44 ? 26 HIS A CA 1
ATOM 3 C C . HIS A 1 1 ? 16.723 2.038 36.064 1.00 18.82 ? 26 HIS A C 1
It seems to me that if we extract the values from the gibberish around them, we can identify the coordiantes for ATOM 1 as:
[ x, y, z]
[18.376, 34.443, 21.34]
To visualise this I'll need to do two things: translate the mmCIF file to coordinates and find a way to display those coordinates
## Extracting coordinates from string soup
I began by
## Things I Learned
- The PDB is a handy open database that contains information on all proteins, including information on what DNA makes them and how they're physically structured.
- FASTA files are proteins in their raw DNA strands. mmCIF files display these proteins folded into position along with a bunch more other data
## Next Time
This section will discuss future improvements and next steps for the project.